DynOmics (dynomics.pitt.edu) is a portal for computing and visualizing biomolecular systems dynamics. At the core of the portal is a newly developed server, ENM 1.0, which permits users to efficiently generate information on the collective dynamics of any structure in PDB format, user-uploaded or database-retrieved.
ENM 1.0 integrates two widely used elastic network models (ENMs)—the Gaussian Network Model (GNM) and the Anisotropic Network Model (ANM), extended to take account of molecular environment. Also, it enables users to assess potentially functional sites, signal transduction or allosteric communication mechanisms, and protein–protein and protein–DNA interaction poses, in addition to delivering ensembles of accessible conformers reconstructed at atomic details based on the global modes of motions predicted by the ANM. The 'environment' is defined in a flexible manner, from lipid bilayer and crystal contacts, to substrate or ligands bound to a protein, or surrounding subunits in a multimeric structure or assembly.
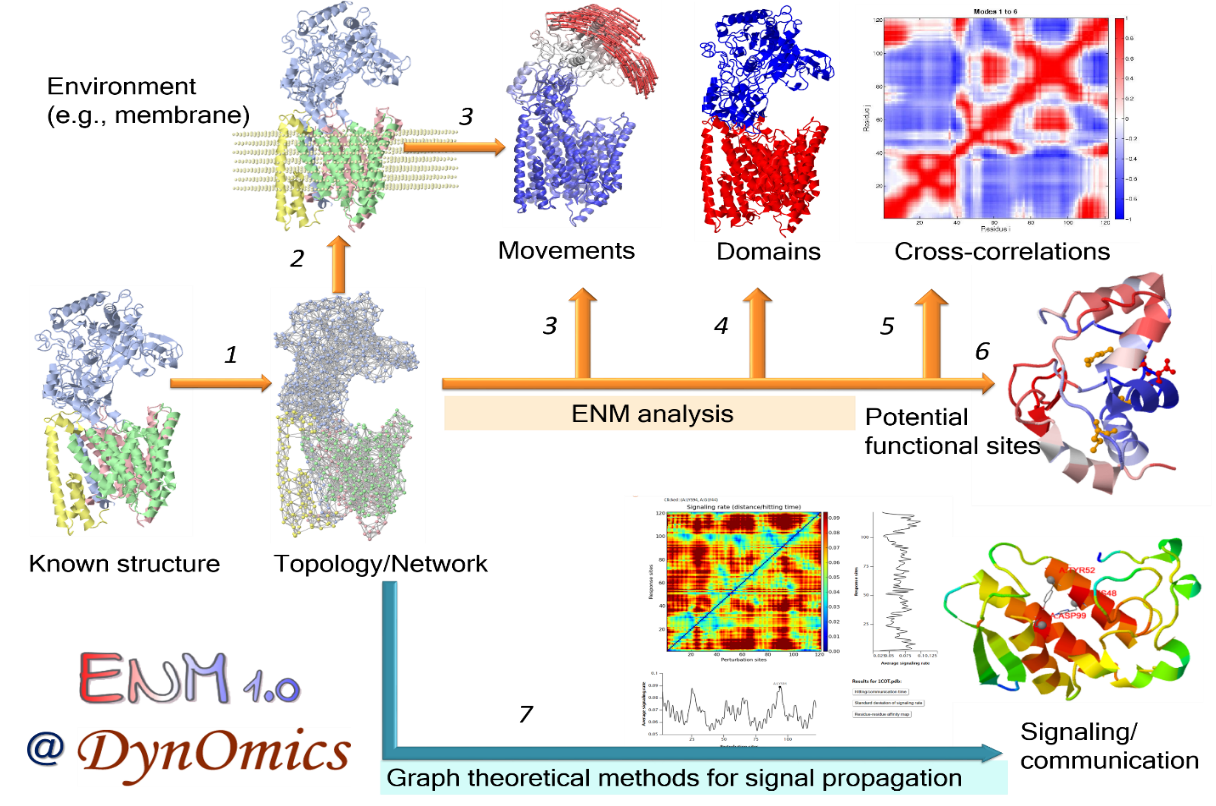
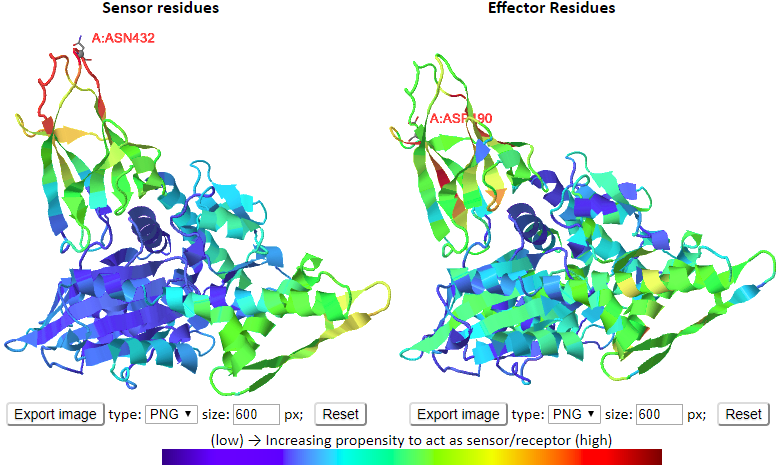
User-friendly interactive features were designed for each component. For examples, users are able to view the predicted sensors and effectors in both 1D/2D profiles and color-coded 3D molecular viewers. And the displaying of 3D molecules are interactive by clicking/labeling in the 1D/2D profiles. It also permits users to easily visualize the movements in the ANM motions and how the environment alter the intrinsic dynamics of the query systems.
|

