|
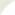 |
|
Allosteric modulation of intact γ-secretase structural dynamics |
Ji Young Lee, Zhiwei Feng, Xiang-Qun Xie, and Ivet Bahar
|
| |
The γ-secretase complex is an attractive target for developing Alzheimer's disease (AD) therapeutics, as abnormal cleavage of the amyloid protein precursor by γ-secretase leads to the aggregation of amyloid-beta (Aβ) fibrils involved in AD. Recent resolution of the 3-dimensional structure of intact γ-secretase opens the way to investigating its structural dynamics in relation to its substrate-binding. γ-secretase has four subunits: presenilin (PS), nicastrin (NCT), anterior pharynx defective 1 (APH-1), and presenilin enhancer 2 (PEN-2). NCT in the extracellular (EC) domain and PS in the transmembrane domain (TMD) have been reported to play important roles in substrate/modulator binding, even though the mechanism of modulation remains to be elucidated.
We analyzed the conformational dynamics of the intact structure in the presence of lipid bilayer using an extension of the anisotropic network model (ANM) that takes account of lipid remodeling. The maps in Fig. 1A-B reveal the engagement of nearby lipid molecules in sustaining the anticorrelated movements of the NCT with respect to the membrane-embedded three subunits of γ-subunit (PS1, APH1, and PEN2). Red blocks indicate correlated substructures; whereas blue regions refer to anticorrelated pairs. The diagrams on right reveal that the NCT acts almost as a lid on PS1 by two dominant types of motions driven by its soft modes: Bending and twisting. Its alternative positions with respect to the PS1 give rise to open or closed conformers that alternately exposes or covers the PS1. Fig 1C shows our druggability results in PS1. Our results detected R1 and R2 sites as druggable, which were already reported to be catalytic site and allosteric site, respectively. Our docking models in Fig 1D provided detailed interactions between ligand and PS1 at R1 and R2, providing deeper insights toward facilitating the structure-based design of novel γ-secretase inhibitor or modulator with therapeutic potential.
Figure 1. ANM dynamics and druggable sites of γ-secretase. (A) Cross-correlation map (left) and corresponding color-coded structure (right) for γ-secretase complex and surrounding lipid bilayer modeled as elastic networks. Results are presented for 10 softest ANM modes. Red blocks along the diagonal indicate strongly correlated substructures, and blue regions indicate anticorrelated (coupled, but moving in opposite direction) pairs of substructures. The color code on the right diagram refers to the correlation of all structural elements (network nodes) with respect to the catalytic residue Asp385 of PS1 (indicated by the yellow-shaded vertical bar on the left map). (B) Close-up view of internal cross-correlations (left) and color-coded diagram (right) as in A, shown for the γ-secretase complex. (C) Two druggable regions of PS1 deduced from druggability simulations performed for the intact protein. Yellow/wheat spheres are druggable hot spots, circled and labeled as R1-R2. (D) Docking of ligands to PS1. Binding of DAPT at the catalytic site is shown on left (identified as R1 in C), and binding of E2012 to the allosteric site is shown on right (identified as R2 in C)
The present study suggests a simple mechanistic model (Fig. 2) for facilitating substrate binding, successive cleavage at two sites, and release of peptide (Aβ) to the EC region, mediated by the structure-encoded global dynamics of the intact γ-secretase complex. We showed that NCT has large bending and twisting motions, allowing for changes in the size of a major groove that has been reported to bind substrate and modulators. Based on the hinge regions observed in our ANM analysis, γ-secretase can be divided into three substructures: TMD (red), NCT small lobe (SL: blue), and NCT large lobe (LL: green). Two hinge regions separate these three substructures, labeled hinge 1 and hinge 2 in Fig. 2A. The NCT fluctuates between open and closed forms, due to the rotational flexibility endowed by these hinges (and thermal fluctuations), either form may be stabilized upon ligand binding. The structure-encoded bending and twisting of the NCT give rise to dramatic changes in the distance between NCT LL and PS1 at the groove; and if the NCT is in the open state (Fig. 2C), the substrate can insert into the groove and bind to the NCT or PS1, but if NCT is closed (as in Fig. 2B), it cannot. NCT motions are thus proposed to play a critical role in allowing or blocking substrate recognition and/or binding. Furthermore, Fig. 2C-D schematically describes the successive ε- and γ-cleavages of the substrate at the two different sites. We propose here that the anticorrelated movements of the NCT to which one end of the substrate is bound may assist in the repositioning of the substrate for ε- and γ-cleavages. Finally, the release of the resulting Aβ-peptide to the EC region can also be facilitated by the opening of the NCT (Fig. 2E).
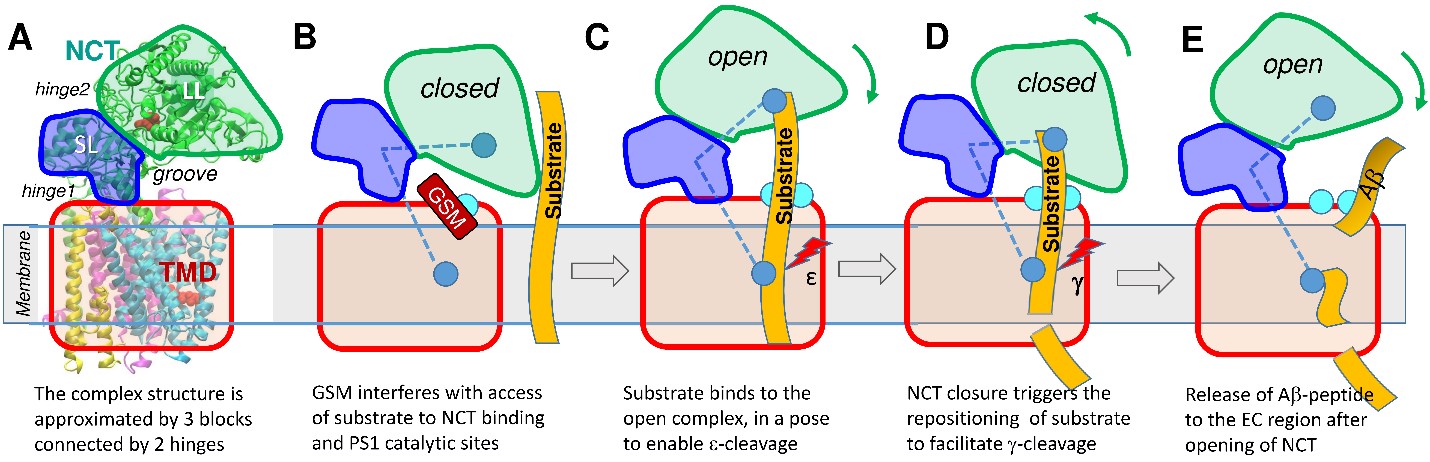
Figure 2. Schematic description of a model for peptide binding, repositioning, cleavage and release modulated by the opening/closure of the NCT with respect the TMD. (A) Schematic diagram of γ-secretase. NCT is composed of two lobes, LL and SL, shown in green and blue separated by hinge2, and the transmembrane domain (TMD) is in red, with interfacial hinge1 enabling the relative movement of NCT SL with respect to TMD. The NCT fluctuates between open and closed states. (B) Binding of modulator (GSM, dark red) to the allosteric site (cyan) near HL1/TM2 (on PS1) or the interlobe groove (on NCT) stabilizes the closed form and obstructs the access of the substrate peptide (orange, extended from EC to TM region) to its binding site and to the PS1 catalytic site. Blue ball in NCT is substrate binding site and blue ball in TMD is catalytic site. (C) Suggested binding pose of substrate, attached on top to the NCT, and exposed at the bottom to the catalytic site. (D) Sliding of the substrate to expose a new cleavage site to the catalytic region, facilitated by the closure of the NCT. Open and closed states may thus help proper positioning of the successive cleavage sites: ε-cleavage occurs in open state C and γ-cleavage, in closed state D. (E) Subsequent opening of NCT permits the release of the Aβ-peptide to the EC region.
|
| |
References
J. Y. Lee, Z. Feng, X.-Q. Xie, and I. Bahar, Allosteric modulation of intact γ-secretase structural dynamics, Biophys. J. Volume 113, Issue 12, p2634–2649, 19 December 2017. |
|
|
|
|
|
|

