|
 |
|
Shared signature dynamics tempered by local fluctuations enables fold adaptability and specificity |
She Zhang, Hongchun Li, James M. Krieger, and Ivet Bahar
|
| |
Recent studies have drawn attention to the evolution of protein dynamics, in addition to sequence and structure-based on the premise structure-encodes-dynamics-encodes-function. Of interest is to understand how functional differentiation is accomplished while maintaining the fold, or how intrinsic dynamics plays out in the evolution of structural variations and functional specificity. We performed a systematic computational analysis of 26,899 proteins belonging to 116 CATH super-families. Characterizing cooperative mechanisms and convergent/divergent features that underlie the shared/differentiated dynamics of family members required a methodology that lends itself to efficient analyses of large ensembles of proteins. We therefore introduced, SignDy, an integrated pipeline for evaluating the signature dynamics of families based on elastic network models, as an addition to ProDy. SignDy can take inputs of protein structures from multiple databases, including PDB, Pfam, CATH, and Dali, and perform structural alignment on a large set of proteins with only low or no sequence similarity. Modes are also calculated and matched to enable comparative studies of their profiles and frequencies, as well as derived dynamical properties such as mean-square fluctuations and covariance matrices.
Our analysis confirmed that family members share conserved, highly cooperative (global) modes of motion. Importantly, our analysis discloses a subset of motions that sharply distinguishes subfamilies, which lie in a low-to-intermediate frequency (LTIF) regime of the mode spectrum. This regime has maximal impact on functional differentiation of families into subfamilies, while being evolutionarily conserved among subfamily members. Notably, the high frequency (HF) end of the spectrum also reveals evolutionary conserved features across and within subfamilies; but in sharp contrast to global motions, HF modes are minimally collective. Modulation of robust/conserved global dynamics by LTIF fluctuations thus emerges as a versatile mechanism ensuring the adaptability of selected folds and the specificity of their subfamilies. SignDy further allows for dynamics-based categorization as a new layer of information relevant to distinctive mechanisms of action of subfamilies, beyond sequence or structural classifications.
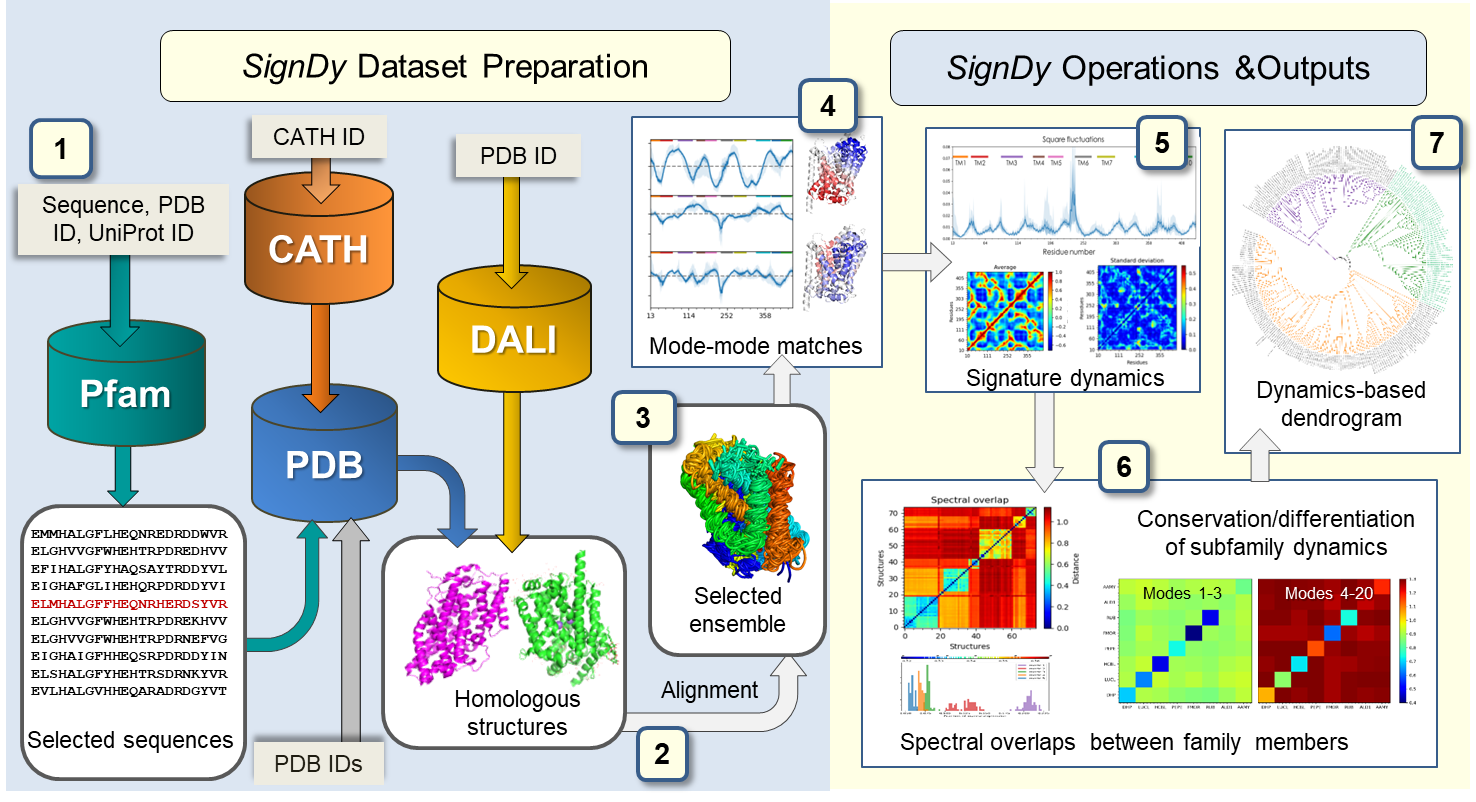
SignDy workflow. The workflow is separated into two main parts: dataset preparation (left) and SignDy operations and outputs (right), described in Zhang et al. MBE (2019). Cylinders and light grey rectangular boxes represent databases and corresponding query inputs, respectively.
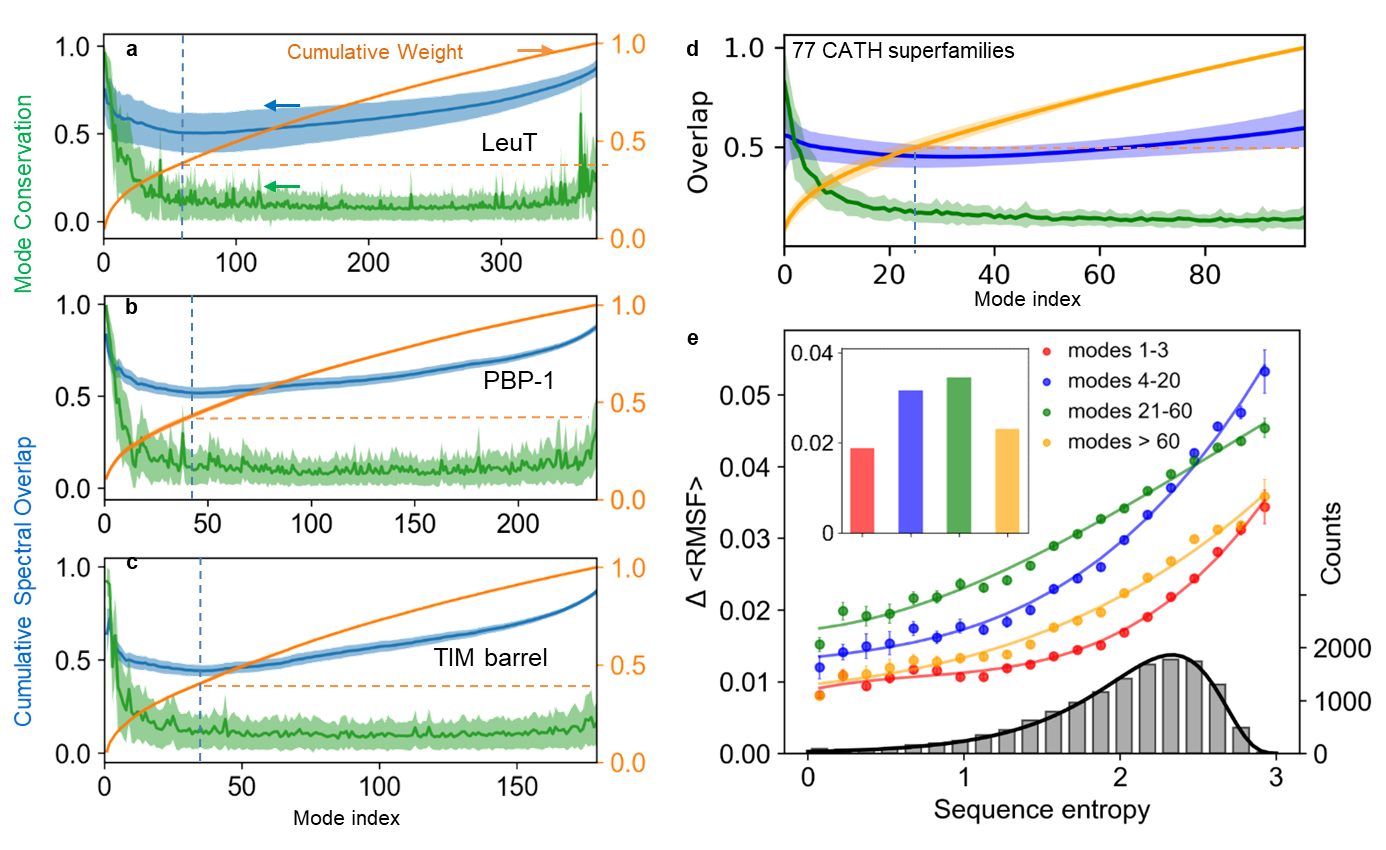
Mode conservation and spectral overlap analysis shows the high conservation of global modes and differentiation of LTIF modes among (super)family members. (a-c) Mode conservation profile given by mode-mode correlation cosines <cc_k> averaged over all family members (green), cumulative spectral overlaps (blue), and cumulative weights of individual modes (orange) plotted as a function of mode index for LeuT, PBP-1 and TIM-barrel folds, respectively. The curves display the averages over all members in each family and the bands show the standard deviations. (d) Same result for first 100 modes obtained for 77 CATH super-families with N > 100. The range 1 = k = 100 covers four regimes of motions: global/softest (k = 3), low frequency (LF) (4 = k = 20), LTIF (21 = k = 60) and high frequency (HF) (k = 60). (e) Change in root-mean-square fluctuations, DRMSF, computed for all residues in each of the 77 CATH super-families as a function of sequence variations (sequence entropy) evaluated for four frequency regimes (labelled). The distribution of sequence entropy for the 77 super-families, shown by the grey bars (right ordinate), fits a lognormal probability distribution (black curve) with a correlation coefficient of 0.997.
|
| |
Related publication:
Zhang S, Li H, Krieger JM, Bahar I. (2019) Shared Signature Dynamics Tempered by Local Fluctuations Enables Fold Adaptability and Specificity. Mol Biol Evol 36: 2053-2068. PMID: 31028708 PMCID: 6736388
Download paper Supplemental Information
Molecular Biology and Evolution, Volume 36, Issue 9
She “John” Zhang, James M. Krieger, and Ivet Bahar are affiliated with the Department of Computational and Systems Biology, University of Pittsburgh School of Medicine, 3501 Fifth Ave, Suite 3064 BST3, Pittsburgh, PA 15213 USA
Hongchun Li, formerly in the Bahar Lab, is now affiliated with the Institute of Biomedicine and Biotechnology in the Shenzhen Institutes of Advanced Technology, the Chinese Academy of Sciences. Guang Dong Province, Shenzhen, PRC.
|
|
|
|
|
|
|

