|
 |
| |
|
|
Huntington's disease (HD) is a devastating chronic neurodegenerative disorder afflicting 30,000 Americans, with 150,000 at risk. HD is caused by a highly penetrant, autosomal dominant mutation in the HTT gene. Expansion of a run of CAG triplets at the 5'-end of this gene increases the number of tandem glutamine residues in the encoded protein (HTT). The connection between glutamine expansion in HTT and neuronal death is not completely understood, no effective treatment for HD neurodegeneration is available and the disease is universally fatal.
We report the implementation of Quantitative Systems Pharmacology (QSP) to Huntington's Disease, with the application of a chemogenomics platform to identify strategies to protect neuronal cells from mutant Huntingtin induced death. QSP is a drug discovery approach that integrates computational and experimental methods in an iterative way to gain a comprehensive, unbiased understanding of disease processes to inform effective therapeutic strategies.1 Chemogenomics is a drug discovery approach that integrates targets and pathways by using active compounds as probes, to characterize the molecular basis of a particular phenotype. Once the hypothesis of the molecular basis is generated, target-based experiments are designed to verify the hypothesis, novel active compounds for further development are identified as well.
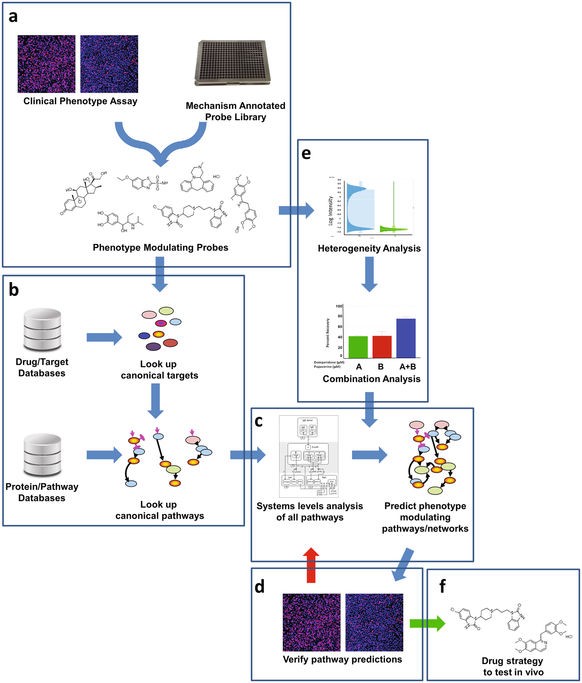
Chemogenomics component of the QSP platform.
Using the STHdhQ111 cell model, we investigated the protective effects of small molecule probes having diverse canonical modes-of-action to infer pathways of neuronal cell protection connected to drug mechanism. Thirty-two mechanistically diverse protective probes were identified, most of which showed less than 50% efficacy. Specific combinations of these probes were synergistic in enhancing efficacy. Computational analysis of these probes revealed a convergence of pathways indicating activation of PKA. The PKA inhibitor H89 at pharmacodynamically active non-toxic concentrations inhibited the effects of several protective compounds, in addition, the integrated intensity of the pCREB signal (a surrogate marker for PKA activity) measured in the nucleus of the STHdhQ111 cells demonstrated the inhibition of PKA activity by H89, thereby supporting the hypothesis that these protective compounds may be working, in part, through activation of the PKA network. The systems-level studies described here can be broadly applied to any discovery strategy involving small molecule modulation of disease phenotype.
|
References
- Stern, A. M.; Schurdak, M. E.; Bahar, I.; et al. A Perspective on Implementing a Quantitative Systems Pharmacology Platform for Drug Discovery and the Advancement of Personalized Medicine. Journal of Biomolecular Screening 2016.
- F. Pei, H. Li, M.J. Henderson, S.A. Titus, A. Jadhav, A. Simeonov, M.C. Cobanoglu, S.H. Mousavi, T. Shun, L. McDermott, P. Iyer, M. Fioravanti, D. Carlisle, R.M. Friedlander, I. Bahar, D.L. Taylor, T.R. Lezon, A.M. Stern, M.E. Schurdak, Connecting Neuronal Cell Protective Pathways and Drug Combinations in a Huntington's Disease Model through the Application of Quantitative Systems Pharmacology, Sci Rep-Uk, 7 (2017) 17803.
|
|
|
|
|
|
|
|
|
|
|

