|
 |
|
Trimerization of dopamine transporter triggered by AIM-100 binding: Underlying molecular mechanisms and effect of mutations |
Mary Hongying Cheng, Luca Ponzoni, Tatiana Sorkina, Jiyoung Lee, She “John” Zhang, Alexander Sorkin, and Ivet Bahar
|
| |
Recent work demonstrated the propensity of dopamine transporters (DATs) to form trimers1, enhanced upon binding a furopyrimidine, AIM-100. AIM-100 binding promotes DAT endocytosis and thereby moderates dopaminergic transmission. Despite the neurobiological significance of these events, the molecular mechanisms that underlie the stabilization of DAT trimer and the key interactions that modulate the trimerization of DAT, and not serotonin transporter SERT, remain unclear. In the present study2, we determined three structural models, termed trimer-W238, -C306 and -Y303, for possible trimerization of DATs using structural data resolved for DAT and its paralogs and structural homologs that share the LeuT fold in advanced computational modeling and simulations. The models are in accord with the versatility of LeuT fold to stabilize dimeric or higher order constructs with selected residues showing a high propensity to occupy interfacial regions. D231-W238 in the extracellular loop EL2, including the intersubunit salt-bridge forming pair D231/D232-R237 (not present in SERT) (in trimer-W238), the loop EL3 (trimers-C306 and -Y303), and W497 on the intracellularly exposed IL5 loop (trimer-C306) and its spatial neighbors (e.g. K525) near the C-terminus are computationally predicted. Site-directed mutagenesis was performed to examine the effect of these elements/sites on DAT oligomerization and endocytosis (enhanced by AIM-100), and the experimental data were further interpreted using a novel machine learning classifier Rhapsody for assessing the impact of single amino acid variants (SAVs) (see Figure 1). The study suggests the possibility of controlling the effective dopamine transport by altering the oligomerization state of DAT upon small molecule binding, as a possible intervention strategy to modulate dopaminergic signaling.
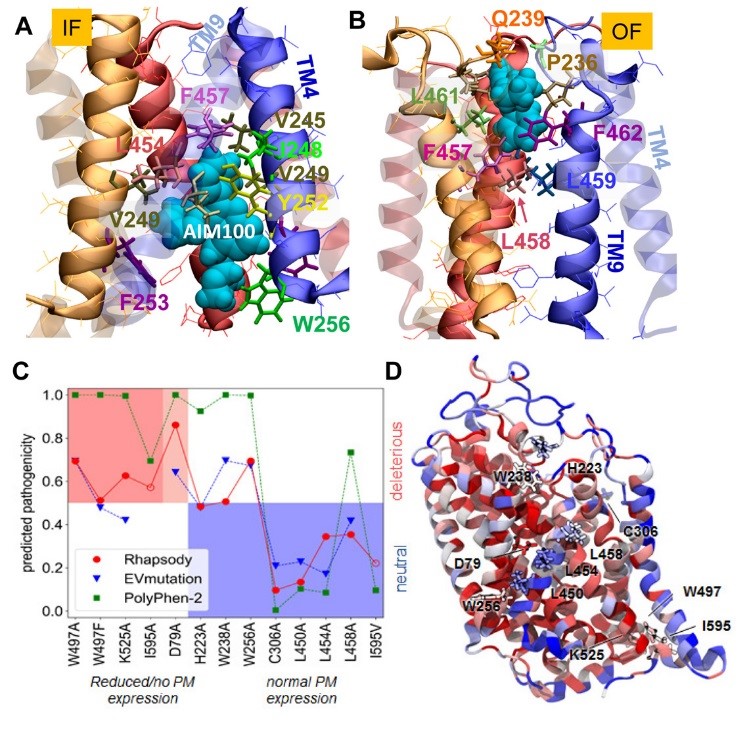
Figure 1: MD simulations reveal the probable binding poses of AIM-100 onto hDAT trimer in the IF (A) and OF (B) state. In silico saturation mutagenesis results and comparison with experimental data. (C) The membrane expression of 13 single amino acid variants (SAVs) has been measured in vitro. On the abscissa, these variants are sorted based on the change in their expression levels (left, red and light-red shade: no or partial expression; right, blue shade: normal/wild-type expression). On the y-axis, the functional impact as predicted by Rhapsody (red circles), EVmutation (blue inverted triangles) and PolyPhen-2 (green squares) is shown. The EVmutation epistatic score has been normalized so that the optimal cutoff between predicted neutral and deleterious effects matches that for Rhapsody and PolyPhen-2 (y = 0.5). Correct predictions lie within the two shaded areas, red (true positives) and blue (true negatives). (D) DAT monomer color-coded by the average pathogenicity shown in C (red/blue: high/low pathogenicity probability). The 13 mutations sites are labelled and shown in licorice.
|
| |
Related publication:
- Sorkina T, Ma S, Larsen M B, Watkins S C, Sorkin A (2018) Small molecule induced oligomerization, clustering and clathrin-independent endocytosis of the dopamine transporter. eLife 7, pii: e32293 PMID: 29630493 PMCID: PMC5896956
- Cheng MH, Ponzoni L, Sorkina T, Lee JY, Zhang S, Sorkin A, and Bahar I. (2019) Trimerization of dopamine transporter triggered by AIM-100 binding: Molecular mechanism and effect of mutations. Neuropharmocology PMID: 31228486 doi: 10.1016/j.neuropharm.2019.107676. [Epub ahead of print]
Supplementary Data:
Download Word document (30MB)
Download zip file
Neuropharmacology, 20 June 2019, 107676
Hongying “Mary” Cheng, Jiyoung Lee, She “John” Zhang, and Ivet Bahar are affiliated with the Department of Computational and Systems Biology, Bahar Lab, University of Pittsburgh School of Medicine, 3501 Fifth Ave, Suite 3064 BST3, Pittsburgh, PA 15213 USA
Tatiana Sorkina and Alexander Sorkin are affiliated with the Department of Cell Biology, Sorkin Lab, University of Pittsburgh School of Medicine 3500 Terrace Street, S362, BST South, Pittsburgh, PA 15261 USA
Luca Ponzoni, formerly in the Bahar Lab, is now affiliated with the University of California, San Francisco in the Keiser Lab, part of the Institute for Neurodegenerative Diseases, the Bakar Computational Health Sciences Institute, the Department of Pharmaceutical Chemistry, and the Department of Bioengineering and Therapeutic Sciences, UCSF Sandler Neurosciences Center, 675 Nelson Rising Lane Room 435, San Francisco, CA 94158
|
|
|
|
|
|
|

