|
 |
 |
In the News...
|
||||||
|
|||||||
 |
 |
 |
|||
|
Congratulatitons to Fank for his PhD graduation, 2025!
Congratulatitons to Mary for her new position as Associate Director, Computational Drug Development at Supernus Pharmaceuticals, Inc. We will miss you, Mary!
Cambridge Workshop: Ivet and students dive into HFSP research, May 30-31, 2024! Engaging talks by Carlos, Satya, and Isabelle + exciting sightseeing!
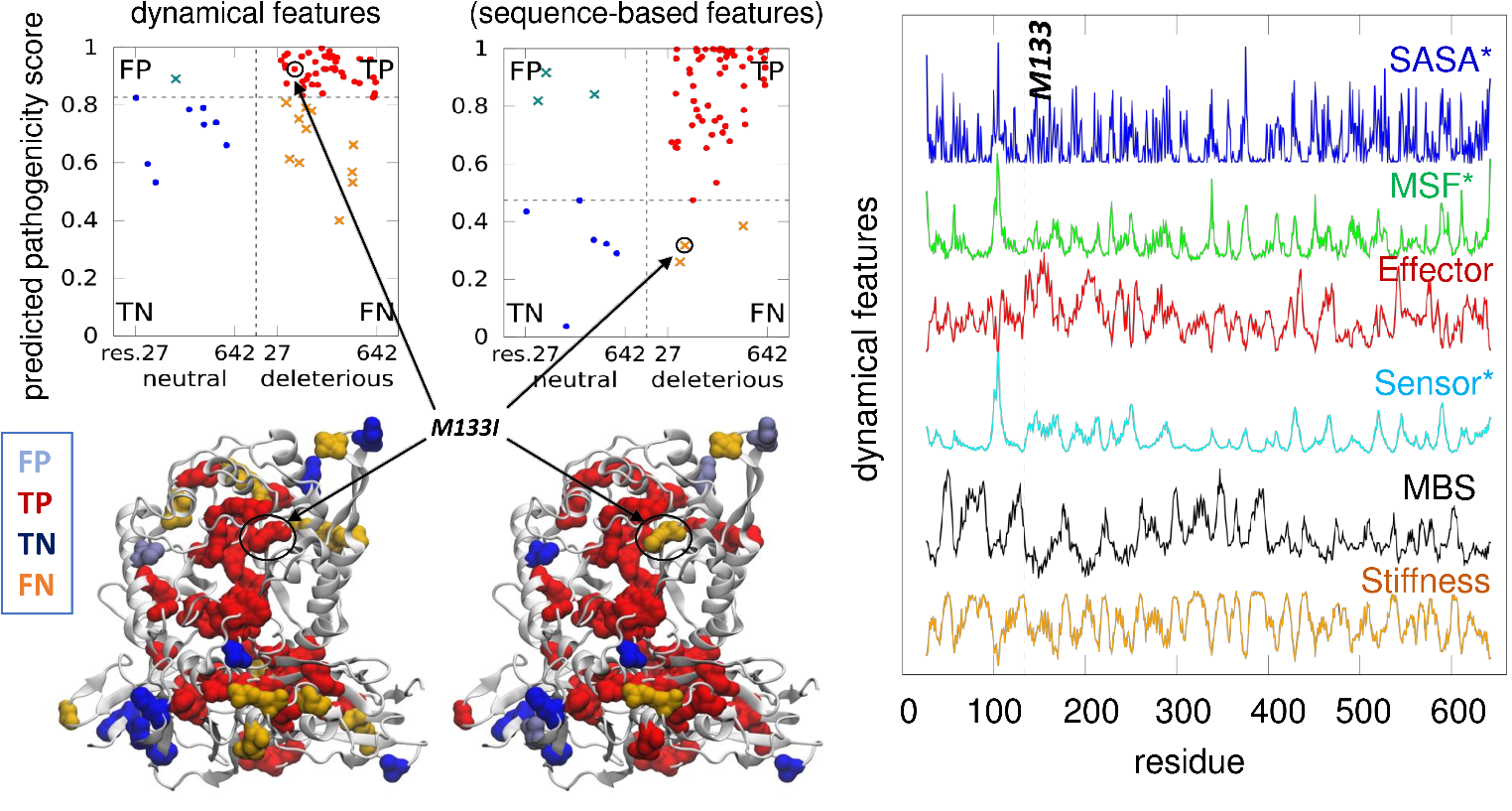
Congratulations to Laufer Center PhD students, Satya, Carlos, and Hoang, for their first contributions to scientific literature: Banerjee, Saha, et al. (2023) Mutually beneficial confluence of structure-based modeling of protein dynamics and machine learning. Curr Opin Struct Biol. 78, 102517; Ventura et al. (2024), Tandem-repeat proteins conformational mechanics are optimized to facilitate functional interactions Curr Opin Struct Biol 84, 10274; Nguyen et al. (2024) Allosteric modulation of serotonin and dopamine transporters Curr Res in Physiol, 100125. This is just the beginning of many more papers to come! Warmest congratulations to Yan for successfully defending his thesis! We wish him the best in his new position at Ilumina.
It is now Hannah's turn to move on to the next stage in her successful career to become an MD. Hannah, it has been such an enjoyable experience to work together!
Fen is off to a successful career as Dr. Pei now, to do postdoctoral research at Silicon Therapeutics, Boston, Massachusetts. Dear Fen, we are so proud, and we all wish you a successful career. We will miss you! Congratulations for TECBio 2018 student Jenea Adams featured in Nature Computational Science for connecting black women in computational biology! We thank her for her kind words: "My main role model was Ivet Bahar [..]. I saw a really powerful woman leading an important department at the university and in computational science, and she inspired me to reach for more and think outside of my comfort zone." It is with mixed feelings that we are saying goodbye to Chelsea for now. It was a pleasure to host her in our lab and we wish her continued success with her studies and her career. We will be here, Chelsea, and still very close while being on different continents. We are certain that our paths will cross again. Thank you again for all of your valuable contributions. Warmest congratulations to John for successfully defending his thesis, and accepting an offer from OpenEye — way to go John! We are so pleased and proud to see that Dr. Hongchun Li is now a Faculty member at Shenzhen Institutes of Advanced Technology, as an Associate Research Professor at the Center for Computer-aided Drug Discovery, Institute of Biomedicine and Biotechnology, Chinese Academy of Sciences. Shenzhen, China. Congratulations, Hongchun, we are so happy for you! |
|
Trimerization of dopamine transporter triggered by AIM-100 binding The Bahar (TR&D1) and Sorkin (DBP3) labs explored the trimerization of dopamine transporter (DAT) triggered by a furopyrimidine, AIM-100, using a combination of computational and biochemical methods, and single-molecule live-cell imaging assays. The study suggests the possibility of controlling dopamine transport upon altering the oligomerization state of DAT by small molecular binding, as a possible intervention strategy to modulate dopaminergic signaling. 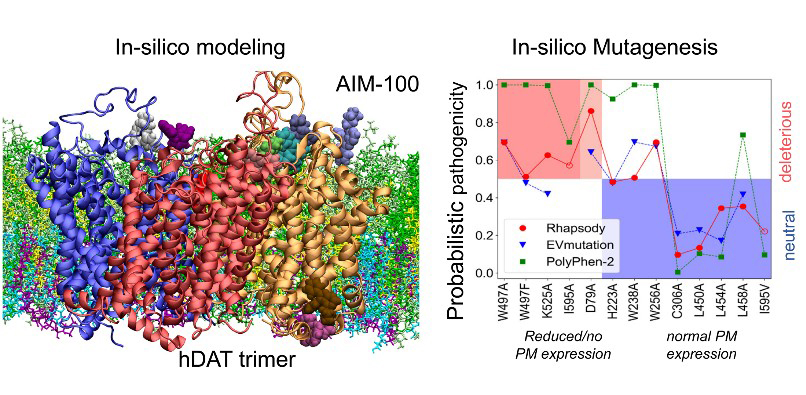 Druggability Simulations and X-ray Crystallography Reveal a Ligand-binding Site in the GluA3 AMPA Receptor N-terminal Domain. We assessed the druggability of the ionotropic glutamate receptor subfamilies, using molecular dynamics simulations in the presence of drug-like molecules and new crystal structures. The study presents a novel ligand-binding site in the GluA3 N-terminal domain and provides pharmacophore features. 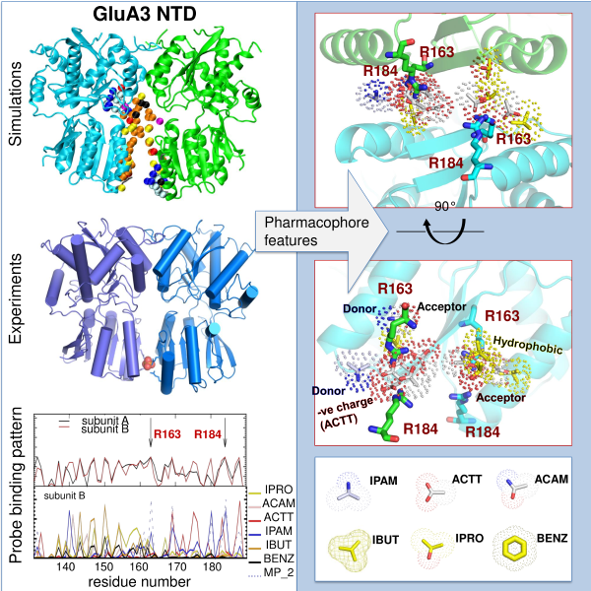 "Systems-level modeling identified a core network that enables cells to 'assess' cellular damage and make a 'life' or 'death' decision upon activating autophagy or apoptosis., in collaboration with the Perlmutter lab. Cytoplasmic Ca2+ acts as a rheostat that fine-tunes autophagic and apoptotic responses through multiple feedback and feedforward loops. The proposed model also provides an in silico platform for developing pharmacological strategies for modulating cell decisions. " 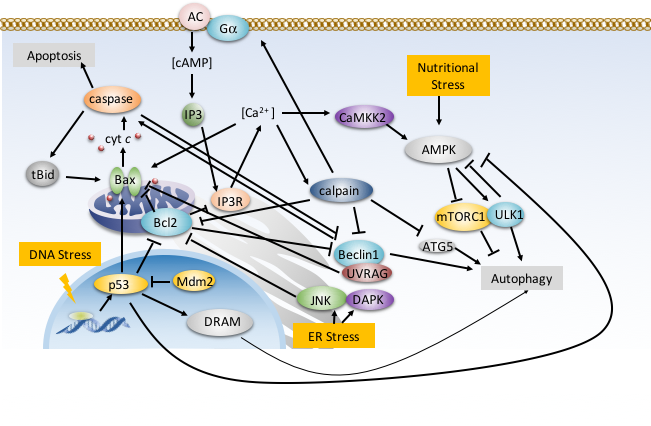
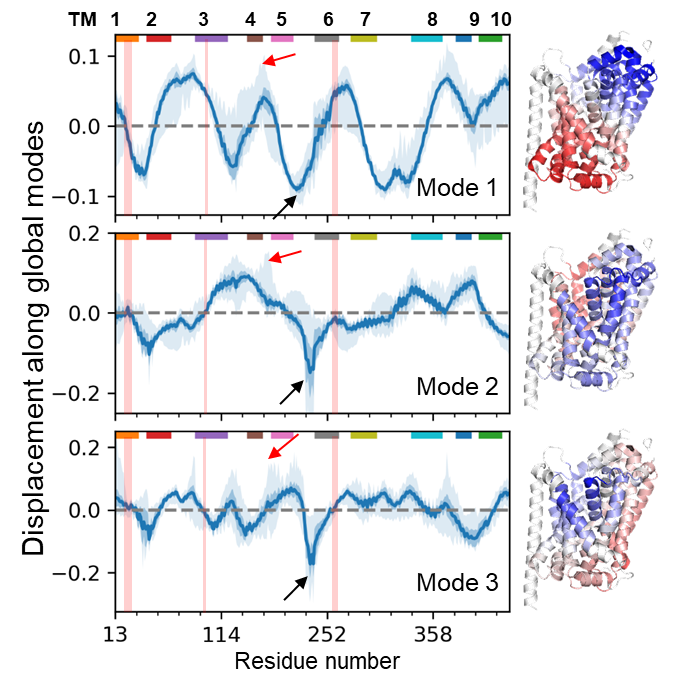
"Allosteric modulation of intact γ-secretase structural dynamics", in collaboration with the Xie lab.
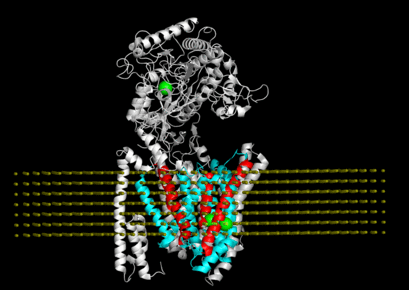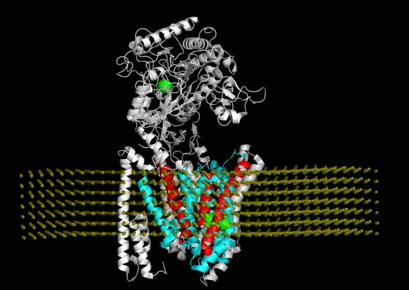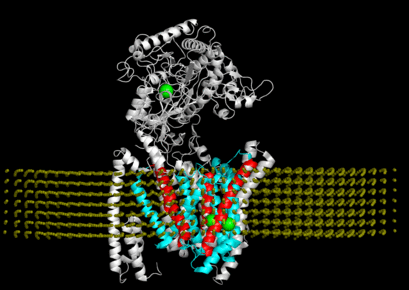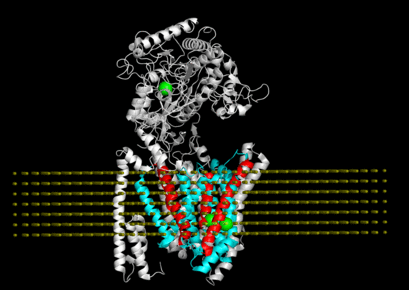 "Connecting neuronal cell protective pathways and drug combinations in a Huntington's Disease model." We implementedQuantitative Systems Pharmacology(QSP) approach to gain a comprehensive, unbiased understanding of Huntington's disease processes to inform effective therapeutic strategies.
Structural dynamics, including allosteric switches, are evolutionarily maintained to accomplish biological activities, consistent with the paradigm sequence -> structure -> dynamics -> function where 'dynamics' bridges structure and function. 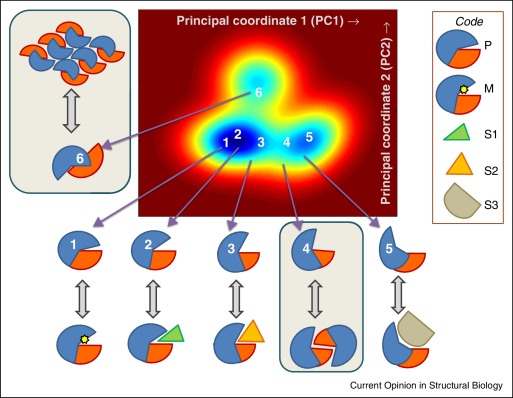
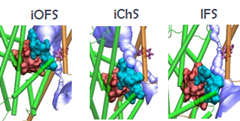
"Adaptability of protein structures to enable functional interactions and evolutionary implications " Turkan Haliloglu and Ivet BaharCurrent Opinion in Structural BIology (2015) 3517-23. Targeting of dopamine transporter to filopodia requires an outward-facing conformation of the transporter. Using quantitative live-cell fluorescence microscopy (Sorkin lab) and molecular modeling (Bahar lab), we investigated the effects of the dopamine transporter (DAT) inhibitor cocaine and its fluorescent analog JHC1-64 on the plasma membrane distribution of wild-type DAT and two DAT mutants. "IFNα2, a Type-I interferon, forms a ternary complex with two receptors, IFNAR1 and IFNAR2. The binding affinity of IFNα2 to these receptors, as well as downstream signaling strength, can be modulated by altering the dynamics of the IFNAR1. IFNα2 associated immune responses were shown to be modulated by introducing Cys-Cys double mutants which form cross-links between different subdomains of IFNAR1. This was a collaborative study with the Schreiber lab at the Weizmann Institute in Israel where experiments (binding and functional assays) have been performed." |
|||